Binary Image Classification of Fungi Species
This project is a proof-of-concept for the task of identifying fungi species from digital images using deep learning. In this preliminary exploration of the idea, I trained a model to correctly classify and differentiate between the Amanita muscaria and Xylodon raduloides species of fungi. The final deployed model performs with an accuracy of roughly 90%. I created a flask application to serve the model on the web. It is hosted on the Google cloud platform and can be accessed at the following link.
 Amanita muscaria
Amanita muscaria
Inspiration
Two biological kingdoms dominate our perception of life in the physical world. In some ways, the fungi kingdom has been overshadowed by plants and animals. Despite their relative obscurity, fungi play an integral role in the Earth’s ecological system and have a remarkably symbiotic relationship with plants and animals, including humans. Among the more than 5 million species of fungi, a diverse array of species contribute to the Earth’s nutrient cycles, influence medical discoveries such as penicillin, and provide nutritional value to humans.
Fascinated by mycology and the relationship of fungi with the natural world, I wanted to apply deep learning methods to create an application that can assist in the identification of species. As over 3000 new species are discovered each year, the taxonomical task becomes quite large. For an untrained eye, it can be difficult to differentiate between certain species that look similar. A misjudgement can have grave consequences, as useful mushrooms often look very similar to poisonous ones. With this project, I try to determine if a learning model can aid in identifying fungi species.
 Xylodon raduloides
Xylodon raduloides
Data
Large datasets of fungi images are relatively hard to find on the internet. Instead of scraping the web to create my own dataset, I used a comprehensive image dataset from the Danish Svamp Atlas. This was the best data I could find, with just over 1300 species with roughly 100 images for each.
The most tedious part of the development process was arguably the data preparation. For this project, I chose two species: Amanita muscaria and Xylodon raduloides. I loaded in the data and performed some key transformations. After resizing, normalizing, and shuffling the data, it was ready to use for training.
Deep Learning
For this project, I used the keras API to create a convolutional neural network model. I used a basic architecture with three convolutional layers with pooling and a final dense layer. For future larger scale versions of this task, I plan to expand to a more powerful architecture. Since this was a simpler binary classification task, I used binary cross entropy for my loss function. So, the model was essentially classifying between 0 and 1.
Model Tuning
While I wanted to keep the model small for this exercise, I still needed to find the right combination of parameters and hyper-parameters for the most optimized network. I used tensorBoard to visualize and compare the performance of various different architectures with different number of convolutional layers, dense layers, kernel sizes, and more.
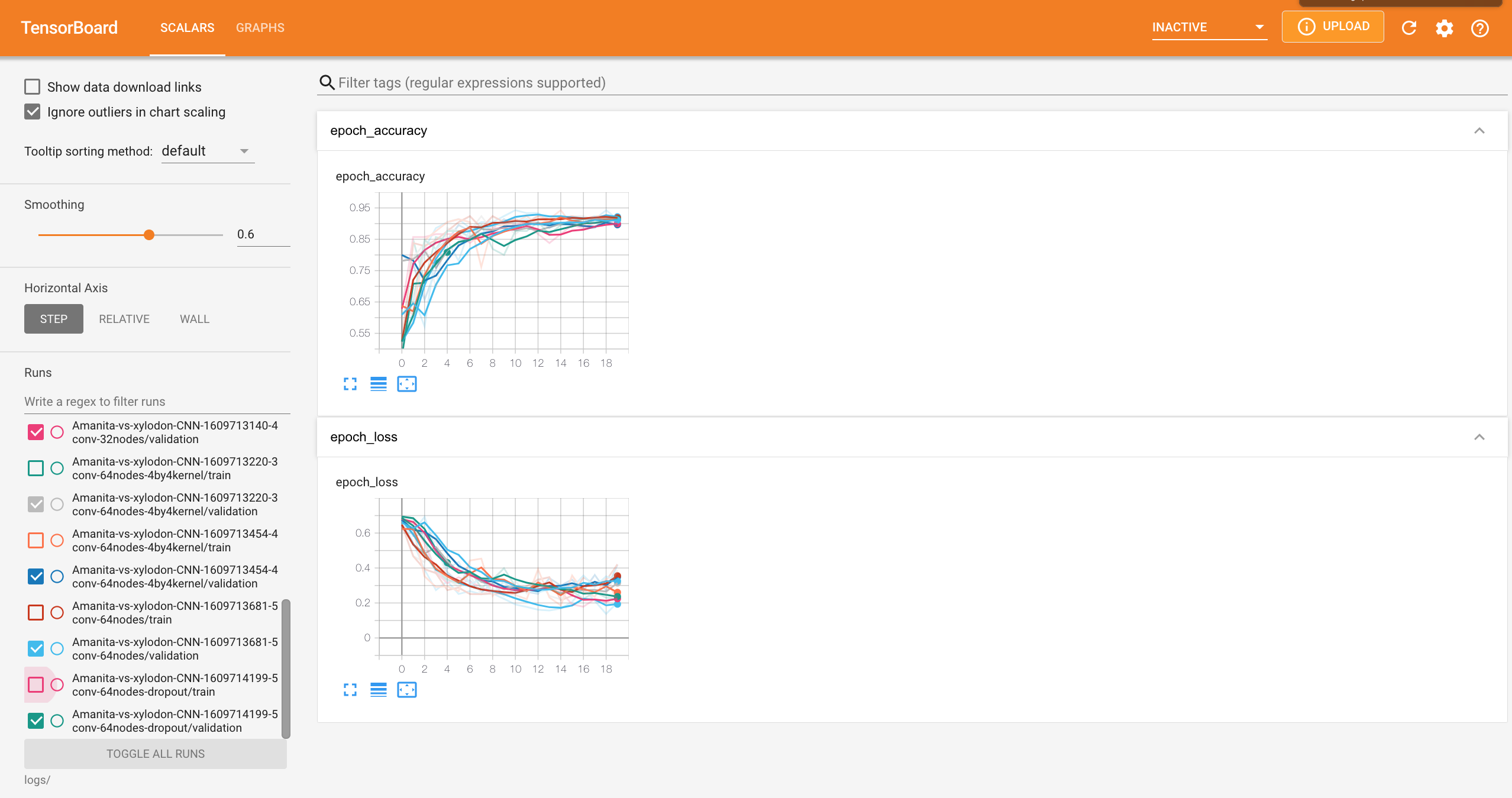
An important note is that I separated my overall data into a training set, a validation set, and a test set. This separation allows us to evaluate our performance based on the model’s ability to generalize the data instead of memorizing it for its inference. In other words, I compared the performance of different models based on the validation accuracy and validation loss.
Web Application
In order to make the model usable to others, I wanted to develop a basic web application that would allow anyone to upload an image of the mushroom they have found and refer to the model to determine the species with some level of confidence.
I used the flask framework to make a simple app with two main actions: uploading an image and predicting the species. I created the webpage with some basic html and css, and saved a model.h5 file with my desired model’s configuration and weights.
Hosting
I used the Google cloud platform to serve the web application. I also used Google cloud storage to store the user-end uploaded images. This was a surprisingly long process, since I had to create and setup the application through Google’s thorough process and develop my app to use specific code to make use of Google cloud storage.
Reflection and Next Steps
The model performs well with outside data it has never seen before. It successfully classified most images from a Google search of the species. There were a few key issues that will need to be addressed in the next stage of this project. The model fails to recognize example images that are unusual from the average appearance of the dataset. For example, the amanita muscaria mushroom is red. If an unusually white amanita mushroom is fed into the model, it will be unsure of the classification. This is an indication that I need more data, through web scraping and data augmentation. Another shortcoming is that the model fails to recognize images of mushrooms from the same genus, but of different species. This is actually a good thing, but it shows that the model may rely on very basic aspects of the image data, such as average color.
For the next stage of the project, my goal is to expand the problem space from just two species to a few dozen, with some species that look similar. This will determine the true potential for deep learning to assist or supersede humans in the classification of fungi species. Future improvements include expanding to multiple classes, data augmentation, more powerful CNN architecture, larger image size, and more training.
As the relationship of fungi with the natural world and its direct effect on us becomes more evident, more of us will begin to explore the largely unexplored landscape of mycology.